
Overview
The principle of DosiTest relies on the assumption that it is possible to model, by Monte-Carlo simulation, a realistic clinical dataset that can be used to benchmark clinical dosimetry procedures. The first step consists in the definition of a realistic clinical model of reference, a virtual patient adapted to molecular radiotherapy.
- A reference anthropomorphic model is used to model patient geometry for the study.
- A reference pharmacokinetics based on literature data is selected.
The reference anthropomorphic model/pharmacokinetics association allows the direct determination of a voxel-based, reference absorbed dose distribution for the study, using Monte Carlo modelling.
For each participating centre, scintigraphic images and associated data are generated at different time points, according to various modalities. As each centre implements a dosimetric approach according to its own criteria, it is essential that each participant receives a simulated dataset that corresponds to what would have been obtained for a patient treated locally.
Each participating centre therefore receives and processes data from the same virtual patient (geometry and pharmacokinetics), but generated specifically to allow the implementation of its own dosimetric protocol. The complete dosimetric study (cumulated activity determination, absorbed dose calculation) is therefore performed locally.
Results and intermediary steps are then forwarded to the centre responsible of the study, to increment a database containing a range of dosimetry results obtained from the same patient, by implementing different clinical dosimetry protocols.
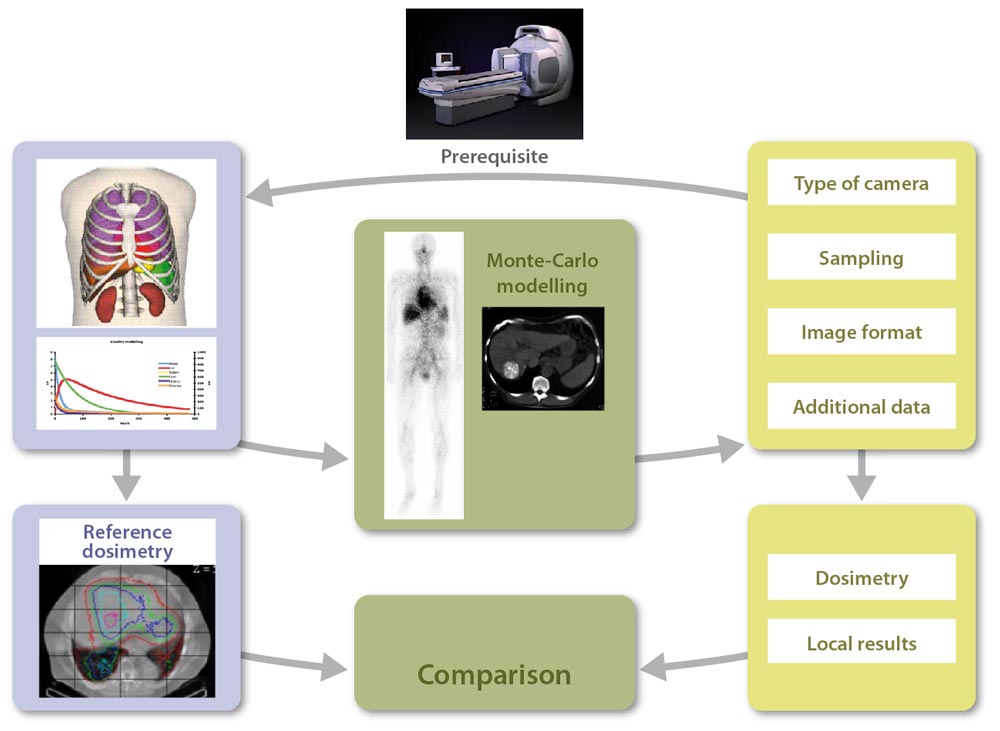
Our lab (hosting lab), creates a virtual patient, a reference dataset {anthropomorphic phantom, pharmacokinetics} defined for all tissues and organs of interest, for a given radiopharmaceutical compound. Monte Carlo modelling is then used for the direct generation of voxel-based reference dosimetry.
Each participating center has to define the data type needed for a dosimetric approach if the study was fully realised locally: planar imaging or SPECT, temporal sampling (number and duration of acquisitions), linked CT images or not, mode of dosimetry calculation, etc.
In addition, they should provide information regarding their gamma camera (type, crystal thickness, etc.), and an example of an experimental image (Jaszczak phantom for example), for us to define their detector model.
We then model the gamma camera used in the participating centre. Modelling validation is done by comparing simulated and experimental images provided by the participating centre. This also allows for testing the import of simulated images in the local image workstations.
Our lab then generates patient images and all additional data required for the dosimetric study (including calibration images), at different times and using a format adapted to the local needs, to obtain a dataset equivalent to what would have been acquired on a real patient.
Scintigraphic images quantification and activity determination are performed by the participating centre, for organs and tissues of interest (liver, kidneys, spleen, lungs, tumours, etc.). The dosimetric study is performed using local resources (commercial or in-house software…).
The participating centre then sends results to the hosting lab to benchmark its approach against the reference. In addition, some intermediate results are transmitted to the hosting lab to allow tracing the different steps of the clinical dosimetry workflow.
The comparison between local results obtained by each participating centre and reference results initially generated by the hosting lab increments a database , thereby allowing the assessment of critical steps in the dosimetry workflow (inducing large variability between centres).
Expected results
By centralizing results and comparing with reference dataset, we will identify critical steps in a dosimetric study. The evaluation of bias introduced by various dosimetric approaches will be performed.
More specifically, we will evaluate the impact of:
- Temporal sampling
- Acquisition mode (planar or tomoscintigraphic)
- Implemented corrections
- Region Of Interest definition (segmentation)
- Background subtraction (in planar mode)
- Absorbed dose determination (pre-computed S factor tables, convolution Monte-Carlo)
Our objective is to participate in the definition of a reference methodology applicable in a clinical context, for a given clinical situation, with associated uncertainties.
The developments required to the realization of DosiTest can be reinvested in a context of virtual scintigraphic imaging, for example to validate nuclear medicine quantitative imaging in silico. This project thus goes beyond the domain of MRT dosimetry.
Finally, DosiTest will generate a unique database that can be used within the context of medical physicist training.
